ENA submission tutorial
The aim of this tutorial is to provide guidance through the process of submitting genomics data to the European Nucleotide Archive (ENA). The task is to submit the genomic sequence data used to create an assembly, to one study, submit the assembly to another study, and then finally group these two studies together under an umbrella study. Throughout the tutorial, the submission steps are exemplified using metadata from submissions done in the Biodiversity Genomics Europe project (BGE).
By the end of this tutorial you will:
- Understand the terminology used by ENA.
- Know how to properly describe and format your genomics data for submission.
- Know how to do a submission.
It serves as complementary material, including examples, to the ENA Documentation.
Preparations
Login page on the ENA submission homepage:
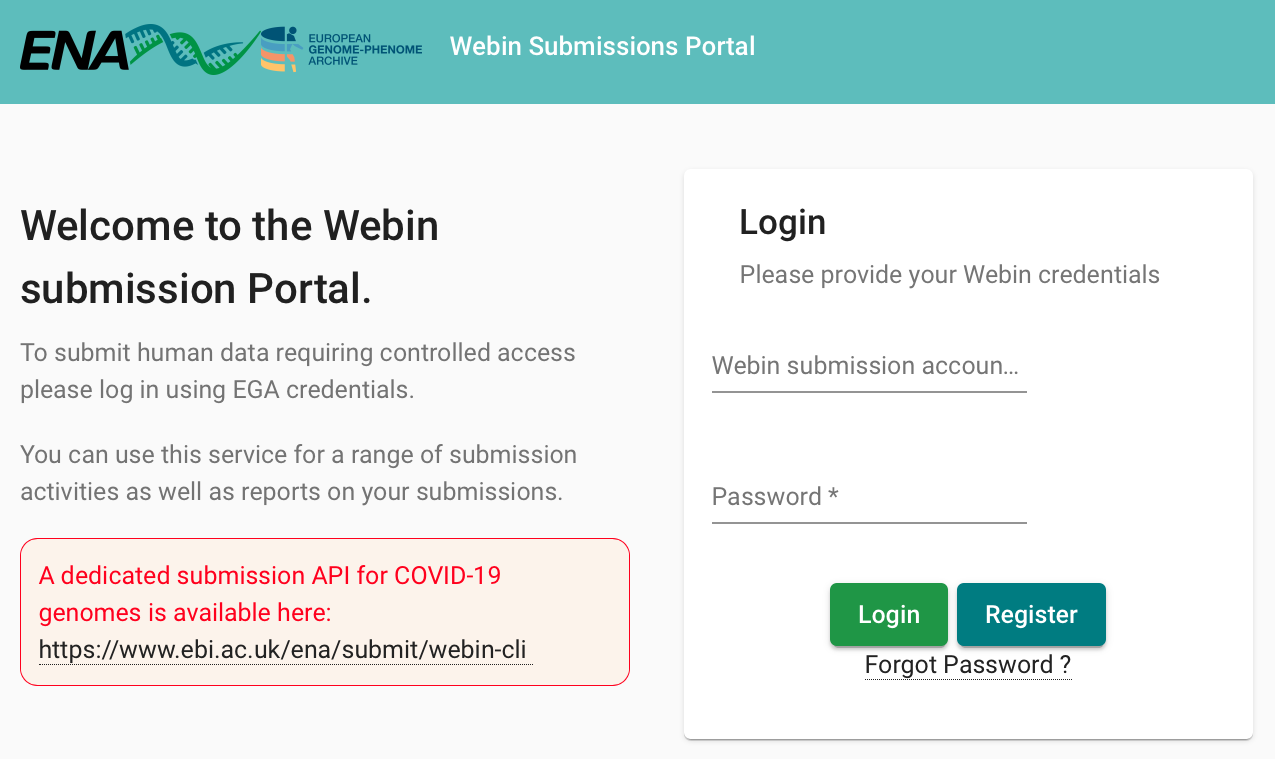
Enter account details to obtain an account in ENA:
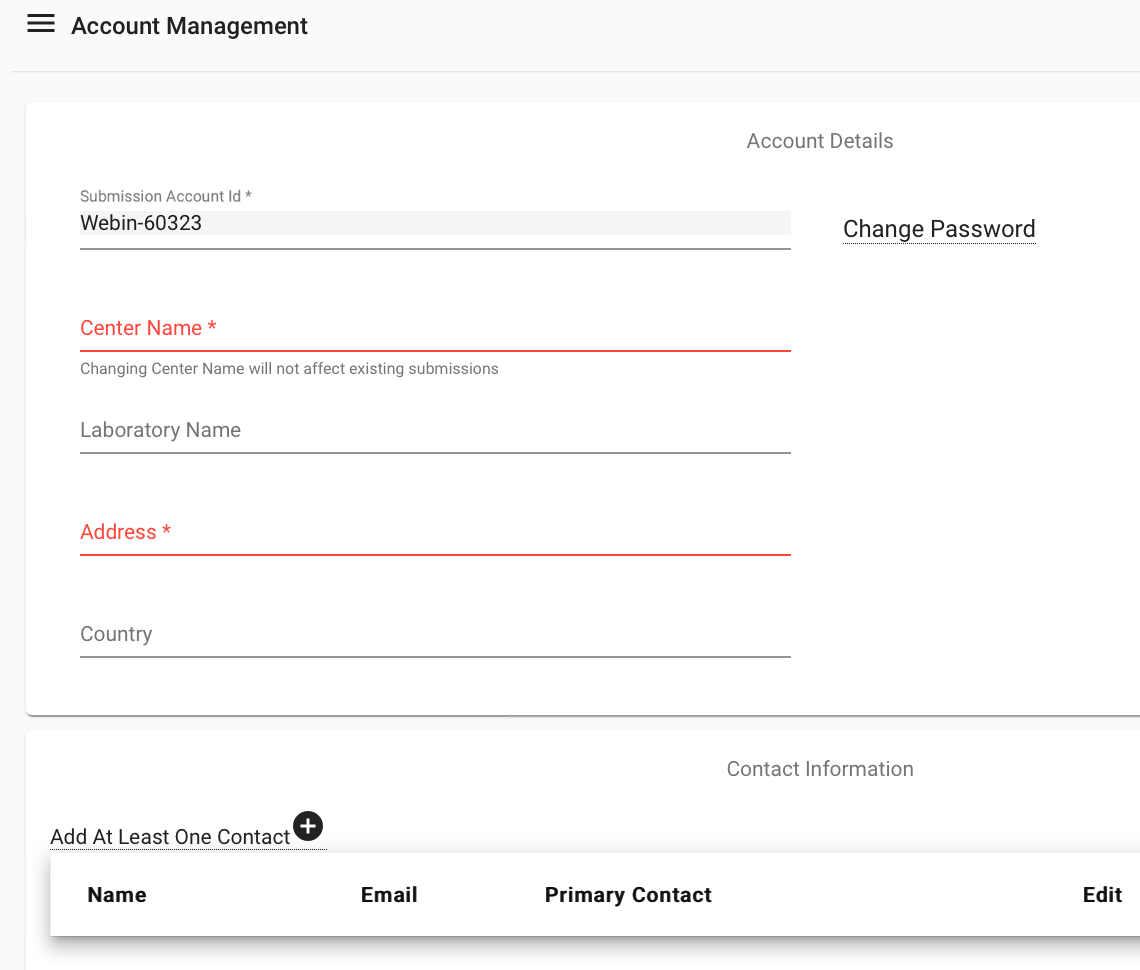
Add contacts
Once you have filled in all of the required fields, you can log in to the submission service. You may wish to add contacts if you are working with other people to complete your submission. You can do this by logging into your account then selecting Home > Manage account. Any contacts that you add will be notified if there are any major changes to data submissions, and they will be listed as contacts on public data.
Download an appropriate template, and fill in the sheets according to the instructions in the template:
- ERC000011 ENA default sample checklist (Excel spreadsheet)
- ERC000015 GSC MIxS human gut (Excel spreadsheet)
- ERC000024 GSC MIxS water (Excel spreadsheet)
- ERC000033 ENA virus pathogen reporting standard checklist (Excel spreadsheet)
- ERC000036 ENA sewage checklist (Excel spreadsheet)
- ERC000050 ENA binned metagenome checklist (Excel spreadsheet)
- ERC000053 Tree of Life Checklist (Excel spreadsheet)
In the sample sheet, the first row contains the following:
- The name of the checklist
- Field names
- Description of the field
- If the field is mandatory, recommended or optional
Note: It is strongly recommended that you provide as much information as possible in the metadata sheets as it will increase the FAIRness of your data, and thus the probability of it being useful in future research efforts.
An example
In this scenario we have used the Tree of life checklist. A completed template is found in Alectoris-graeca-metadata.xlsx, adapted from a real submission made under the Biodiversity Genomics Europe (BGE) project organised by the European Reference Genome Atlas (ERGA) initiative. The task is to submit four datasets (one PacBio HiFi WGS, two Illumina HiC, and one Illumina RNAseq), including four samples, all in one study, plus a chromosome level assembly in another study, and then group these them studies together under an umbrella study. Please note that it is not necessary to submit the assembly in a study separate from the raw reads. In our example it is done to demonstrate how umbrella studies can be used.
Checksums
Typically you will receive the checksums from the data producer in the delivery, but in case you need to calculate them yourself:- Open a terminal (command prompt) window and change to the directory where the raw reads are located, e.g.
cd my_data/raw/. - Modify and execute the command
md5sum * > checksums-md5.txtto match your file names and required output name. The command will calculate the checksums for all files in current directory and list them in a file named checksums-md5.txt.
Data transfers
There are multiple ways of transferring the raw read files to ENA upload area, depending on which type of machine the transfer is initiated from, see ENA documentation on file upload. If all else fails, a simplelftp will work, but large file transfers always risk transfer interruption, and you will have to start all over again. In order to avoid this, it might be worth the effort of installing and using Aspera, since it has the functionality of resuming transfers. For more information regarding using Aspera, please see our topic page on Data transfer.
Submission methods
ENA describes three methods of submission, none of which can be used to complete all parts of a submission: interactive, command line (using Webin-CLI), and programmatic.
Interactive Submission Method
This method involves filling out web forms directly in the browser and downloading template spreadsheets that can be completed off-line and later uploaded to ENA. This is the easiest method to use if you are unfamiliar with submissions, but quickly becomes time-consuming with bulk submission (> 50 records). It is possible to register and submit studies, samples, and raw reads this way.
Test submission
ENA provides two sites for submission; one for test submissions and one for production submissions (i.e. an actual submission to ENA). We recommend doing a test submission first, using the example data provided, in order to get an understanding of the different steps. Note: The test service is purged every 24 h, meaning that any submitted test file will be removed before the following day, making it impossible to begin a submission one day and continue the next.
- ENA's documentation on Register a study interactively.
- Go to ENA login (test) and login.
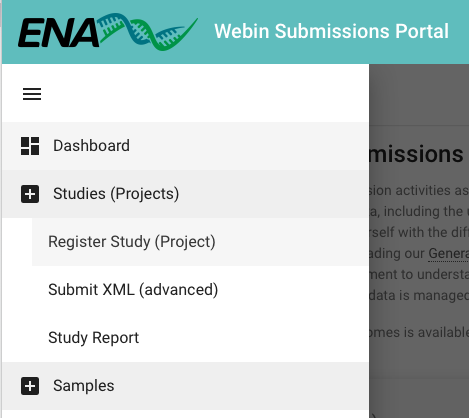
- After logging in, you will see the landing page (shown below) that includes multiple options for completing your registration.
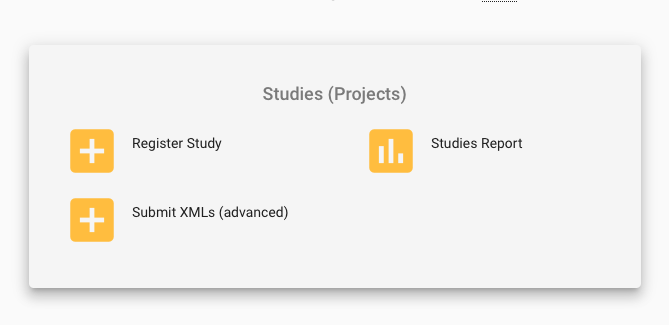
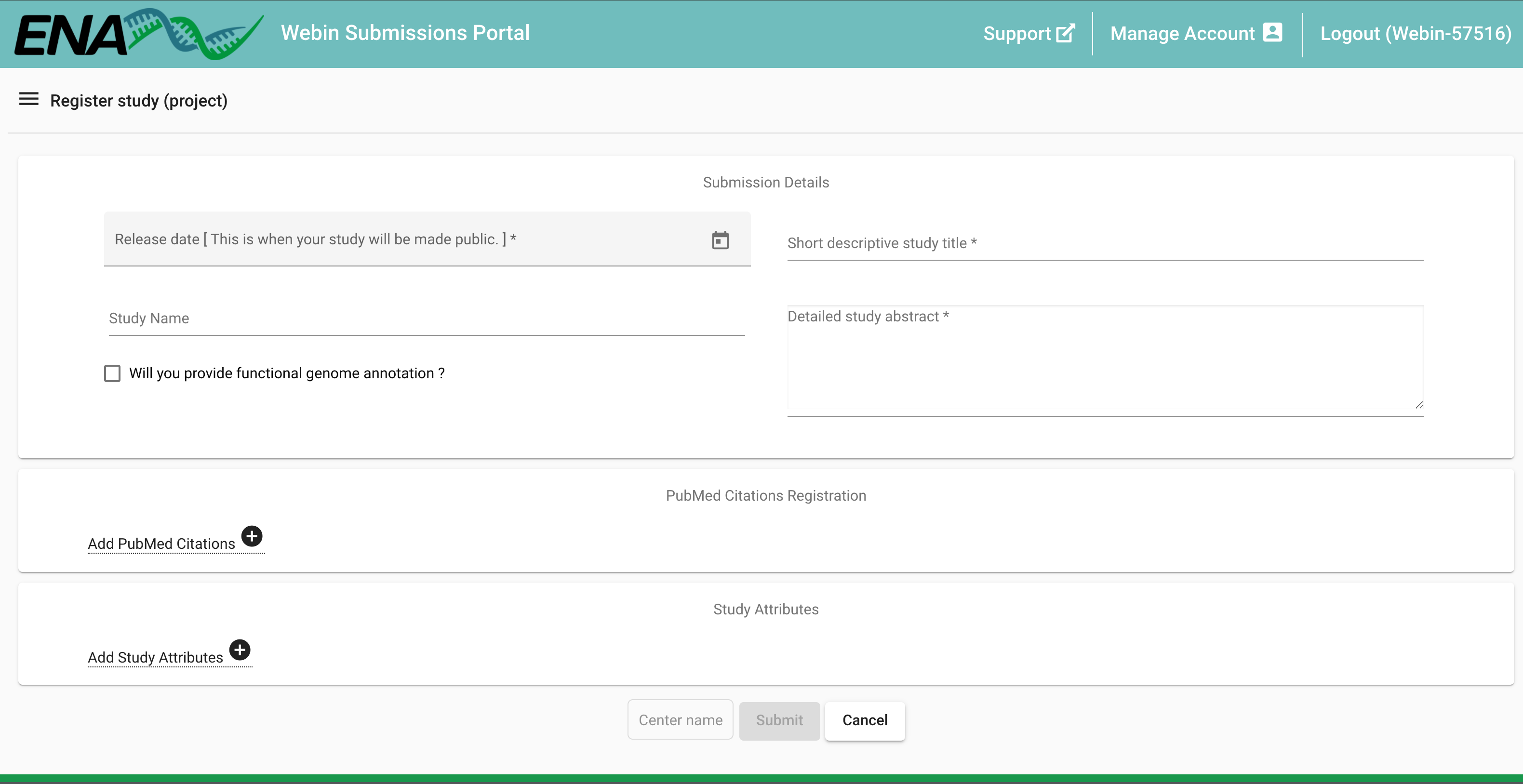
- In the top left of the landing page, there is a Dashboard menu that expands when you click on it. Click it and select Register Study (Project) or click on the Register Study option in the Studies (Projects) section on the landing page (the options for 'Study' are shown in yellow).
- Enter the details of the project, by copy and paste from the metadata template. Asterisks (*) denote mandatory fields. The 'Release date' is the date that the record will become publicly available. This can be updated later, so if you are unsure on a precise date, you can provide an estimated date (maximum two years forward in time). You will get a notification email from ENA, a few weeks prior to the release date, when your data is about to become public. If necessary, the release date can then be consecutively extended any number of times by up to two years.
- In our example the study registration is repeated twice, once for the genomic sequencing data and once for the assembly data. The umbrella study needs to be registered using a different method, see further Umbrella submission in Programmatic Submission Method below.
- Note: In case you have an annotated assembly you must also provide a locus tag field, e.g. with an abbreviation of the scientific name of the species.
- Click Submit. A pop-up window will appear, check that the submission was successful by reading the content of the window, copy the project accession number (starting with PRJEB...) and add it to your metadata template for future reference, then click Close.
- ENA's documentation on Register samples interactively.
- Create Alectoris-graeca-samples.tsv by copying the filled rows of the
ENA_samplessheet of a metadata template in .tsv format. This can be done in any text editor, e.g. Notepad or Visual Studio Code. - Remove rows 3 (with field descriptions) and 4 (with info if mandatory/recommended/optional) from the .tsv file.
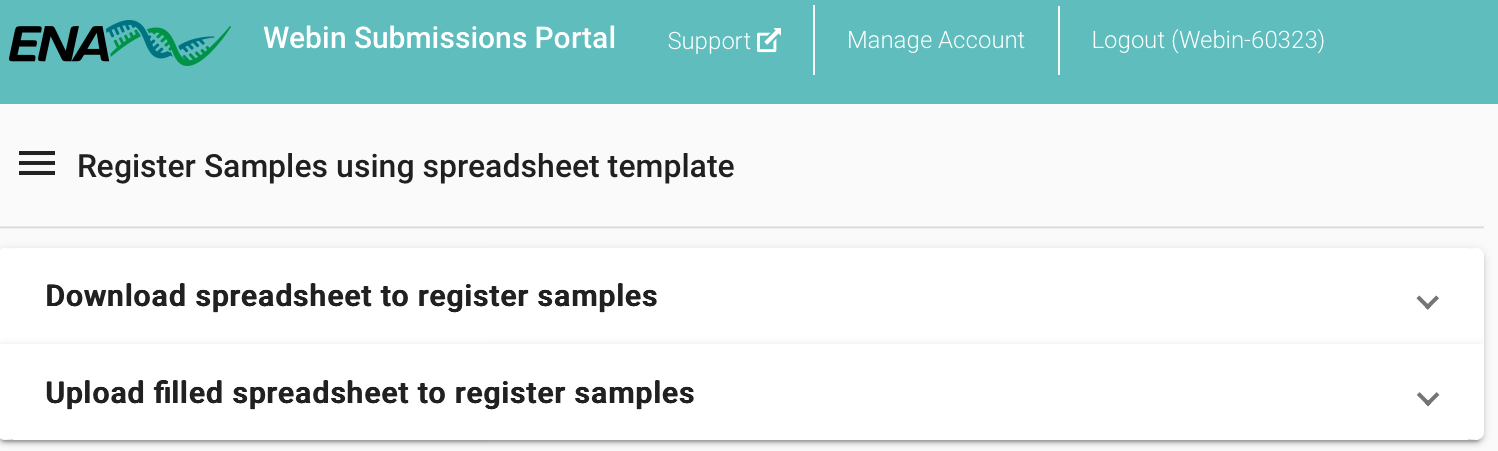
- Go to the browser where you are logged in to ENA, and register the samples either by clicking on the Dashboard menu (top left of the page) and selecting Register Samples or by clicking on the Register Samples option in the Samples section of the landing page (all related options in green).
Both of the above options lead to the same place, which gives two options: (1) Download spreadsheet to register samples, and (2) Upload filled spreadsheet to register samples. - Select the latter and upload the samples .tsv file. Click on Submit Completed Spreadsheet, verify that the submission was successful in the pop-up Submission window, and then click Close.
- ENA's documentation on Submit raw reads interactively.
- Raw reads are submitted in a similar fashion as samples, with the exception of one additional step:
- For interactive submission of raw reads, one needs to combine the information concerning the experiment and the runs into one .tsv file. Hence, copy cells A1:F7 in the 'ENA_run' sheet and paste them to cells added to the cells M2:R8 in 'ENA_experiment' sheet (this is already done in the example .xlsx file).
- Create Alectoris-graeca-experiments.tsv by copying the filled rows of the
ENA_experimentsheet of a metadata template in .tsv format. This can be done in any text editor, e.g. Notepad or Visual Studio Code. - Remove rows 3 (with field descriptions) and 4 (with info if mandatory/recommended/optional) from the .tsv file.
- Go to the browser where you are logged in to ENA, and register the experiments either by clicking on the Dashboard menu (top left of the page) and selecting Submit Reads, or by clicking on the Submit Reads option in the Raw Reads (Experiments and Runs) section of the landing page (all related options in orange).
- Both of the above options lead to the same place, which gives two options: (1) Download spreadsheet template for Read submission, and (2) Upload filled spreadsheet template for Read submission.
- Select the latter and upload the experiments .tsv file. Click on Submit Completed Spreadsheet, verify that the submission was successful in the pop-up Submission window, and then click Close.
Webin-CLI Submission Method
This method uses ENA’s Webin-CLI program. Submissions require the preparation of text (manifest) files that are validated before submissions are completed. It is possible to submit raw reads and assemblies, this way. Actually, for assemblies, this is the only way to submit.
- ENA's documentation on Submitting Genome Assemblies of Individuals or Cultured Isolates.
- The only way to submit assemblies is by using the tool called Webin-CLI, hence the first step is to download the latest version, see ENA's documentation on Webin-CLI Submission. This tutorial will use Java jar, but a Docker image is also available.
- Webin-CLI requires that the metadata is formatted in a manifest file, hence create Alectories-graeca-manifest.txt.
- Note that in order to submit a chromosome level assembly, a gzipped file listing the chromosomes is also needed. An example of what they look like is in chromosome_list.txt. We will also submit a unlocalised_list.txt, which contains a list of unlocalised sequences.
- Webin-CLI can be used in two steps, first to validate and, upon successful validation, to submit:
java -jar ~/webin-cli-8.2.0.jar -ascp -context genome -userName Webin-XXXXX -password 'YYYYY' -manifest ./Alectoris-graeca-manifest.txt -validate -testjava -jar ~/webin-cli-8.2.0.jar -ascp -context genome -userName Webin-XXXXX -password 'YYYYY' -manifest ./Alectoris-graeca-manifest.txt -submit -test - Note: The
-testindicates that the submission will be made to the test instance. Omit this when doing real submissions. - Upon a successful submission, an accession starting with ERZ will be obtained. However, this is not the official accession to be used in a publication. ENA will need to process the assembly, and when this is done you will receive an email with the genome assembly accession to be used (GCA_xxxxxx), e.g.:
ASSEMBLY_NAME | ASSEMBLY_ACC | STUDY_ID | SAMPLE_ID | CONTIG_ACC | SCAFFOLD_ACC | CHROMOSOME_ACC bAleGra1.1 | GCA_965278835 | PRJEB79727 | ERS17759205 | CBDIHD010000001-CBDIHD010000076 | | OZ257071-OZ257110
Programmatic Submission Method
This method requires the preparation of XML files that are sent to ENA using cURL (or uploading them via the Webin Portal of ENA). It is possible to register and submit studies, samples, and raw reads this way. This is the only way to register umbrella studies.
Any programmatic submission requires two .xml files, one with the action (ADD, MODIFY, RELEASE, CANCEL) and one with the metadata. The submission is done via a cURL command in a terminal window, and all levels (study, sample, raw reads) can be registered and submitted at the same time, but in this tutorial the levels will be submitted separately.
Test submission
ENA provides two sites for submission; one for test submissions (https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/) and one for ‘real’ submissions (i.e. an actual submission to ENA) (https://www.ebi.ac.uk/ena/submit/drop-box/submit/). We recommend doing a test submission first, using the example data provided, in order to get an understanding of the different steps. Note: The test service is purged every 24 h, meaning that any submitted test file will be removed before the following day, making it impossible to begin a submission one day and continue the next.
- ENA's documentation on Register a study programmatically.
- Create submission.xml, with the action, and update the desired release date. It can be set to maximum two years forward in time, and be adjusted later on. You will get a notification email from ENA, a few weeks prior to the release date, when your data is about to become public.
- Create Alectoris-graeca-study.xml, containing the metadata, with the names, titles, and descriptions of the raw reads study and the assembly study. Note that there is an
aliasadded to each of the studies,bAleGra-study-raw-readsandbAleGra1-study-assembly. These are used in the submission of experiment and assembly, respectively, to be able to refer to which study they should be submitted to. This is handy if you want to submit all levels of metadata in the same cURL command, since you wouldn't know the accession number of the project at that point. - Submit in a terminal window by typing:
curl -u username:password -F "SUBMISSION=@submission.xml" -F "PROJECT=@Alectoris-graeca-study.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/" - A receipt will be written in the window, with
success="true"orsuccess="false". If true you will get the accession numbers, note these down in the .xlsx file. An example receipt:<?xml version="1.0" encoding="UTF-8"?> <?xml-stylesheet type="text/xsl" href="receipt.xsl"?> <RECEIPT receiptDate="2024-09-05T08:24:23.589+01:00" submissionFile="submission.xml" success="true"> <PROJECT accession="PRJEB79726" alias="bAleGra-study-raw-reads" status="PRIVATE" holdUntilDate="2025-05-10Z"> <EXT_ID accession="ERP163841" type="study"/> </PROJECT> <PROJECT accession="PRJEB79727" alias="bAleGra1-study-assembly" status="PRIVATE" holdUntilDate="2025-05-10Z"> <EXT_ID accession="ERP163842" type="study"/> </PROJECT> <SUBMISSION accession="ERA30781837" alias="SUBMISSION-05-09-2024-08:24:23:241"/> <MESSAGES> <INFO>All objects in this submission are set to private status (HOLD).</INFO> </MESSAGES> <ACTIONS>ADD</ACTIONS> <ACTIONS>HOLD</ACTIONS> </RECEIPT>
- ENA documentation on Register samples programmatically.
- Create Alectoris-graeca-samples.xml, with the metadata for the four samples.
- As with the study, a submission.xml is needed. We can reuse the one already created, but the HOLD action will not have any effect since all samples are private 'forever' unless an experiment refers to it. The sample will become public the same time as the experiment becomes public.
- Register in a terminal window by typing:
curl -u username:password -F "SUBMISSION=@submission.xml" -F "SAMPLE=@Alectoris-graeca-samples.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/" - Upon success of the submission, each sample will obtain a BioSample accession (starting with SAMEA) and one internal ENA accession (starting with ERS).
- ENA's documentation on Submit raw reads programmatically.
- Two metadata .xml files are needed in order to submit raw reads, one each for the experiments and the runs.
- Create Alectoris-graeca-experiments.xml and Alectoris-graeca-runs.xml
- A note on
alias. We have created aliases for both studies and samples, which is not necessary if one submits each metadata object separately, because thenrefname=""can be replaced byaccession=""and the accessions received in previous steps can be added. However, when it comes to submission of raw reads, we will submit both objects at the same time and thus we don't yet know what the experiment accessions will be. For the experiment aliases, we will use a combination of the sample alias and the library name, and the run aliases are named with the experiment alias and the respective file type as an addition at the end. - As in the previous step, the submission.xml created for the study will be reused also here.
- Submit in a terminal window by typing:
curl -u username:password -F "SUBMISSION=@submission.xml" -F "EXPERIMENT=@Alectoris-graeca-experiments.xml" -F "RUN=@Alectoris-graeca-runs.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/" - Upon success of the submission, both experiments and runs will obtain accession numbers(starting with ERX and ERR, respectively). The run accessions will be used in the next step, assembly submission.
- ENA's documentation on Create an Umbrella Study.
- Create a submission-umbrella.xml, with the ADD action and an umbrella.xml where the two studies (one for the raw reads and one for the assembly) are added as children.
- Note: Umbrella studies are automatically created with a release date two years ahead in time. A separate submission, with the action RELEASE, is done when it is time to make the umbrella public, see how in ENA's documentation Releasing Umbrella Studies.
- Register in a terminal window by typing:
curl -u Username:Password -F "SUBMISSION=@submission.xml" -F "PROJECT=@umbrella.xml" "https://wwwdev.ebi.ac.uk/ena/submit/drop-box/submit/" - Umbrella studies cannot be seen when you login to your account. Hence, it is even more important that you make sure that the term
success="true"is present in the typed receipt, and that you note down the received accession number in your metadata .xlsx file.
Terminology
- 'Raw' sequence data - Sequence data that is obtained directly from a sequencing instrument (typically in FASTQ or BAM file format).
- Analysed sequence data - Sequence data that has been processed in some way after being obtained from a sequencing instrument. Such data has been normalised, and perhaps also subject to other processing (e.g. removal of outliers, calculation of expression measurements, and statistical analyses). Another example of analysed data is genome assemblies.
- Metadata - Description of the data that gives, at a minimum, sufficient information to reproduce the data collection method (e.g. description of how the source material was obtained and details about the sequencing process, such as library preparation and instruments used).
- Study - A study (project) object is used to group together all data submitted to ENA about a given study and to control its release date. A study accession number is typically used to cite data submitted to ENA. Note that all data and metadata associated with a study are made public together with the study when it is released.
- Sample - A sample object contains information about the sequenced source material. Checklists are used to define which fields should be filled when describing samples. Note that a taxonomic classification system is used to refer to biological organisms; the accepted organism name and classification hierarchy are used, see The European Nucleotide Archive (ENA) taxonomy for further details.
- Experiment - An experiment object contains all the details about the metholodology used for sequencing, including library preparation protocols used and instrument details.
- Run - A run object is part of an experiment object. It refers to data files that contain the raw reads.
- Analysis - An analysis object contains secondary analysis results. These results are derived from the raw reads (e.g. a genome assembly).
Resources
Please find below resources concerning ENA submission tutorial - in form of training, guidance and/or tools.