Sharing
In the era of FAIR (Findable, Accessible, Interoperable and Reusable) and Open science, datasets should be made available to the public. Whenever possible, discipline-specific repositories (with or without controlled access) should be used in order to increase the FAIRness of your research outputs. If no such repository is available, there are general-purpose repositories.
Learn more about the FAIR principles
Finding a suitable repository type
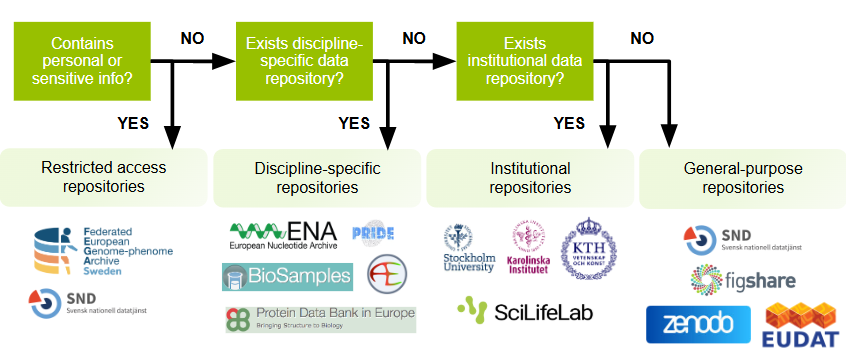
Does your data contain personal or sensitive information that cannot be fully anonymised?
There may be cases where openly sharing data is not feasible due to ethical or confidentiality considerations. Depending on what the ethical board approving your study said about data sharing, and the level of permission granted from participants, it may still be possible to make your data accessible to authenticated users via a controlled-access repository.
See more guidelines on sharing human data
Is there a discipline specific repository for your dataset?
Research data differs greatly across disciplines. Discipline-specific repositories offer specialist domain knowledge and curation expertise for particular data types. Using a discipline-specific repository makes your data visible to others in your community.
Does your institutional repository accept data?
Many institutions offer support to their employees for managing and depositing data. Institutional repositories that accept datasets provide stewardship, helping to ensure that your dataset is preserved and accessible.
Answered no on all questions above?
General-purpose data repositories accept datasets regardless of discipline or institution. These repositories support a wide variety of file types and are particularly useful where a discipline-specific repository does not exist.
Recommended discipline-specific repositories
There are several repositories for life science data types (see e.g. ELIXIR Deposition Database list), please find a selection of them below.
Genomics data
Click on the buttons below for specific information regarding suitable repositories for sharing genomics data.
We also recommend the following two specialised guidelines:
- Guide for submission of metabarcoding data to ENA provided by the Swedish Biodiversity Data Infrastructure (SBDI)
- Tutorial for SARS-CoV-2 genome data submission to ENA provided by the Swedish Pathogens Portal
For convenience, we have created templates for the most frequent data types and their corresponding ENA checklists. The templates come with instructions on how to do an interactive submission, via the ENA Webin Portal, but even when doing a programmatic submission, the template can be useful for collecting all necessary descriptions / metadata. Download an appropriate template, and fill in the sheets according to the instructions in the template:
- ERC000011 ENA default sample checklist (Excel spreadsheet)
- ERC000015 GSC MIxS human gut (Excel spreadsheet)
- ERC000024 GSC MIxS water (Excel spreadsheet)
- ERC000033 ENA virus pathogen reporting standard checklist (Excel spreadsheet)
- ERC000036 ENA sewage checklist (Excel spreadsheet)
- ERC000050 ENA binned metagenome checklist (Excel spreadsheet)
- ERC000053 Tree of Life Checklist (Excel spreadsheet)
Visit ENA
Visit ArrayExpress
Visit EGA
Visit FEGA Sweden
Imaging data
Depending on the type of image data you have, different public repositories are available, please see the table at BioImage Archive.
Visit BioImage Archive
Metabolomics data
MetaboLights is a database for Metabolomics experiments and derived information. The database is cross-species, cross-technique and covers metabolite structures and their reference spectra as well as their biological roles, locations and concentrations, and experimental data from metabolic experiments.
Visit MetaboLights
Proteomics data
The ProteomeXchange Consortium provides globally coordinated standard data submission and dissemination pipelines involving the main proteomics repositories.
Visit PRIDE
Visit PeptideAtlas
Other data
Guidance on where to publish COVID-19 and Pandemic Preparedness research data, can be found on the Swedish Pathogens Portal.
For other discipline-specific repositories, see e.g. ELIXIR Deposition databases, EBI archive wizard (help to find the right repository depending on data type), or FAIRsharing (the latter can also assist in finding metadata standards suitable for describing your datasets).
Institutional repositories
For datasets that do not fit into discipline-specific repositories, it is recommended to use an institutional repository if available. These repositories are often general-purpose repositories with the intention of only being used by researchers at that specific institution. See below a selection of available repositories offered by Swedish academic institutions operating in the life sciences.
Visit KI Data Repository
Visit KTH Data Repository
Visit KTH Zenodo community
Visit SciLifeLab Data Repository
Visit Stockholm University Figshare
General-purpose data repositories
A general-purpose data repository is an appropriate choice only if the data does not need to be published in a controlled-access repository, a discipline-specific repository does not exist for the discipline and if there are no institutional repositories available. See below a selection of general-purpose data repositories.
Visit DORIS
Visit Figshare
Visit Zenodo
Visit EUDAT B2SHARE
How can SciLifeLab help you sharing data?
If you are a researcher at a Swedish academic institution operating in the life sciences, you can get help from SciLifeLab Data Management support team. This support team can help you describe and deposit your data. Here are a few examples of the support that is offered:
- Plan data submission
- Identify suitable repositories
- Assist during the submission process when publishing data and code
- Assist in the creation of metadata records in SciLifeLab Data Repository
- Advice on what needs to be done when working with sensitive human data
- Advice on describing data with proper metadata
If you need any help connected to data submission, please contact us!
Resources
Please find below resources concerning the research data life cycle phase share in form of training, guidance and/or tools.
Training resources
Guiding resources
- AIDA Data Sharing Policy
- Annotare - ArrayExpress submission tool
- Data with personal information in DORIS
- EBI submission YouTube playlist
- European Nucleotide Archive (ENA) user documentation
- Guide for submission of metabarcoding data to ENA
- Handbook for data containing personal information
- How to submit data to PRIDE
- Keynote: First FAIR steps, Rob Hooft
- National guidelines for open science
- NBIS Submission documentation
- RDMkit on Data organisation
- RDMkit on Data publication
- RDMkit on Documentation and metadata
- RDMkit on Sharing data
- Research Involving Human Data by Richelle Björvang
- SciLifeLab Data Repository Metadata record tutorial
Tools
- AIDA Data Hub
- Annotare - ArrayExpress submission tool
- ArrayExpress
- Aspera CLI install instructions
- BioImage Archive
- BioStudies
- DORIS
- EBI repository wizard
- ELIXIR Deposition Databases for Biomolecular Data
- ENA checklists
- EUDAT - B2SHARE
- European Genome-phenome Archive (EGA)
- European Nucleotide Archive (ENA)
- FAIRsharing
- Federated European Genome-phenome Archive Sweden
- Figshare
- KI Data Repository
- KTH Data Repository
- MetaboLights
- PASSEL - PeptideAtlas submission tool
- PeptideAtlas
- PRIDE Submission Tool
- PRoteomics IDEntifications Database (PRIDE)
- protocols.io
- Rclone documentation
- Registry of Research Data Repositories (re3data)
- Researchdata.se
- SciLifeLab Data Repository
- Stockholm University Figshare
- The Swedish Reference Genome Portal
- Zenodo